AQuaRef: Machine Learning Accelerated Quantum Refinement of Protein Structures
Our collaborative work on AQuaRef is now published in Nature Communications. This AI-enabled quantum refinement method leverages AIMNet2 to achieve unprecedented accuracy in protein structure determination from cryo-EM and X-ray crystallography data.
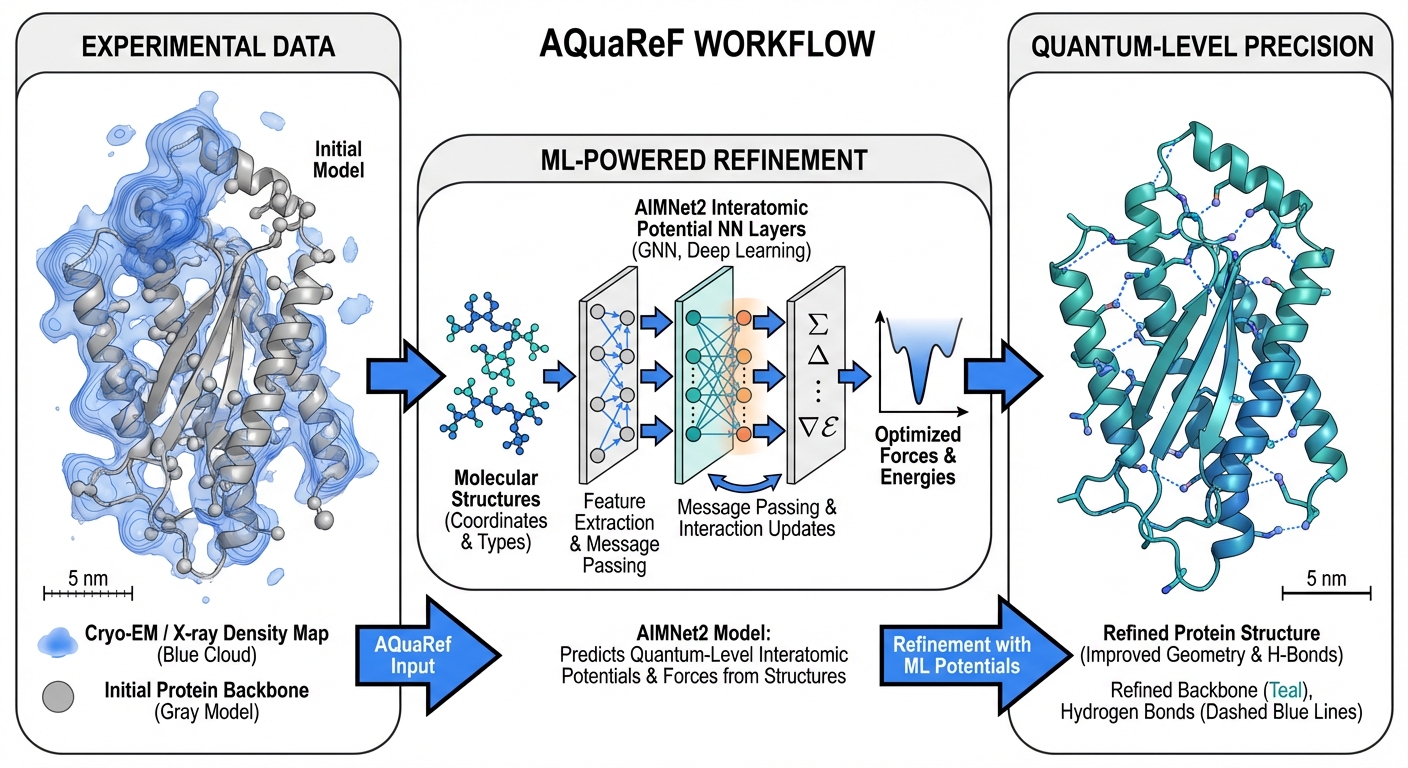
We are pleased to announce the publication of our collaborative work on AQuaRef in Nature Communications. This research represents a significant advance in structural biology, demonstrating how machine learning interatomic potentials can revolutionize the refinement of protein structures obtained from cryo-electron microscopy (cryo-EM) and X-ray crystallography experiments.
The Challenge of Protein Structure Refinement
Cryo-EM and X-ray crystallography provide the experimental data essential for determining the three-dimensional structures of proteins and other biomacromolecules at atomic resolution. However, the refinement process—converting raw experimental data into accurate atomic models—relies heavily on library-based stereochemical restraints. These libraries, while useful, have two fundamental limitations: they are restricted to known chemical entities, and they do not adequately capture meaningful noncovalent interactions that govern protein folding and function.
Quantum mechanical (QM) calculations offer a theoretically superior alternative, as they can describe all chemical interactions from first principles without requiring pre-defined restraints. Unfortunately, the computational cost of QM methods scales prohibitively with system size, rendering them impractical for the large molecular systems typically encountered in structural biology.
AQuaRef: Bridging the Gap with Machine Learning
AQuaRef (AI-enabled Quantum Refinement) addresses this challenge by leveraging AIMNet2, a machine learned interatomic potential that mimics QM accuracy at a fraction of the computational cost. The method integrates seamlessly with Phenix, a widely used software suite for macromolecular structure determination, enabling researchers to apply quantum-level refinement to their structures without requiring specialized expertise in quantum chemistry.
The approach was validated through extensive benchmarking on 41 cryo-EM and 30 X-ray crystallographic structures. The results demonstrate that AQuaRef consistently yields atomic models with superior geometric quality compared to conventional refinement techniques, while maintaining an equal or better fit to the experimental data. Critically, the method achieves this improvement without the overfitting issues that can plague other approaches.
Determining Proton Positions in Challenging Cases
One of the most notable capabilities of AQuaRef is its ability to accurately determine hydrogen atom positions—a task that is notoriously difficult with conventional methods because hydrogen atoms scatter X-rays and electrons weakly compared to heavier atoms. The paper illustrates this capability through the challenging case of short hydrogen bonds in the parkinsonism-associated human protein DJ-1 and its bacterial homolog YajL.
Short hydrogen bonds are of particular biological interest because they often occur at enzyme active sites and play critical roles in catalysis. Accurate determination of proton positions in these bonds is essential for understanding reaction mechanisms but has historically required neutron diffraction experiments, which are far more resource-intensive than X-ray or cryo-EM studies. AQuaRef demonstrates that machine learning can provide this information directly from standard crystallographic or cryo-EM data.
Collaborative Effort Across Institutions
This work represents a collaborative effort spanning multiple institutions, including Carnegie Mellon University, Lawrence Berkeley National Laboratory, University of Florida, University of Wrocław, and Pending.AI. The interdisciplinary team brought together expertise in machine learning, quantum chemistry, and structural biology to create a tool that addresses a real need in the structural biology community.
The integration of AIMNet2 with Phenix exemplifies how foundational advances in machine learning potentials can be translated into practical tools that benefit researchers across disciplines. AIMNet2’s ability to handle diverse chemical environments—including charged species and element-organic compounds—makes it particularly well-suited for the varied chemistry encountered in protein structures containing cofactors, ligands, and post-translational modifications.
Implications for Structural Biology
The implications of this work extend beyond improved geometry scores. More accurate atomic models lead to better understanding of protein function, more reliable drug design, and more precise mechanistic insights. As cryo-EM resolution continues to improve and the technique becomes increasingly central to structural biology, methods like AQuaRef that can extract maximum information from experimental data will become ever more valuable.
Furthermore, because AQuaRef operates within the familiar Phenix environment, adoption barriers are minimal. Structural biologists can enhance their refinement workflows without learning new software or investing in specialized computational resources.
Read the full paper: AQuaRef: machine learning accelerated quantum refinement of protein structures — Roman Zubatyuk, Malgorzata Biczysko, Kavindri Ranasinghe, Nigel W. Moriarty, Hatice Gokcan, Holger Kruse, Billy K. Poon, Paul D. Adams, Mark P. Waller, Adrian E. Roitberg, Olexandr Isayev, and Pavel V. Afonine, Nature Communications, 2025, 16, 10.1038/s41467-025-64313-1.
We are grateful to our collaborators and funding agencies for making this work possible, and we look forward to seeing how the structural biology community applies AQuaRef to advance our understanding of protein structure and function.